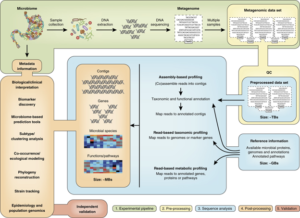
Metagenome sequencing is transforming pathogen surveillance.
Relevance:
- GS Paper 3 Health
Recent Update:
One of the initial breakthroughs in the definitive identification of SARS-CoV-2 as the causative agent of COVID-19 came from the application of unbiased genome sequencing technologies to infected patient samples. This new approach – called metagenomics. The technique and its adoption also drastically changed the way pathogen identification would be undertaken thereafter.
What’s in todays article?
- What is ‘metagenomics’.
- What is ‘genomics’.
- key findings of Nigerian study.
- Potential of metagenomics in public health.
Metagenomics:
- Metagenomics is the study of the structure and function of entire nucleotide sequences isolated and analyzed from all the organisms (typically microbes) in a bulk sample.
- Metagenomics is often used to study a specific community of microorganisms, such as those residing on human skin, in the soil or in a water sample.
- The broad field may also be referred to as environmental genomics, eco-genomics, community genomics or micro-biomics.
Genomic tech to the frontline:
- Scientists in countries worldwide have since developed scores of technologies based on genome-sequencing – including the very popular CovidSeq assay – spawning several national and international SARS-CoV-2 genome surveillance activities.
- GISAID, a popular repository on the Internet to which SARS-Co-V-2 genome-sequence data could be submitted, is a testimony to such high-throughput genome surveillance activities.
- India also initiated a national SARS-CoV-2 genome-sequencing and surveillance program supplemented by several state government and private initiatives.
- The success has provided a template for applying sophisticated genomic technologies as frontline tools to surveil known and unknown organisms and to tracking emerging pathogens in an unbiased and high-throughput manner.
The Nigerian study:
- In a paper in Nature Communications, scientists from the Nigerian Centre for Disease Control applied metagenomic sequencing for pathogen surveillance and detection in three cohorts of patients.
- The first cohort represented population-level surveillance of individuals presenting with symptoms consistent with Lassa fever, a viral haemorrhagic fever caused by the Lassa virus endemic to West African countries.
- The second cohort consisted of people from outbreaks with suspected infectious aetiologies.
- The third cohort consisted of people with clinically challenging but undiagnosed conditions.
- The scientists were able to identify 13 distinct viruses afflicting the individuals. The genomic approach also helped them pinpoint the second and third documented cases of human blood-associated dicistrovirus among the cohorts.
- The scientists also identified pegivirus C as a common co-infection in patients with Lassa fever as well as that the presence of pegivirus C was associated with a lower viral load in patients.
- As it happened, the metagenome-sequencing approach also allowed the scientists to rule out viral infections in some individuals and link their symptoms to pesticide poisoning instead.
- The study demonstrated the power of metagenomic sequencing investigations for pathogen detection and disease diagnosis, and to inform public health outbreak responses.
Key to early response:
- More recently, experts have used genome sequencing technologies as frontline tools to motivate the detection and surveillance of lumpy skin disease in cattle and the emergence of drug-resistant tuberculosis, among the use-cases.
- It is heartening that several initiatives worldwide are taking advantage of the speed, accuracy, and high-throughput nature of advanced genome sequencing technologies to detect pathogens from diverse environmental sources, such as wastewater, air, soil, and animals.
What is genomics?
- Genomics is the study of the total or part of the genetic or epigenetic sequence information of organisms, and attempts to understand the structure and function of these sequences and of downstream biological products.
- Genomics in health examines the molecular mechanisms and the interplay of this molecular information and health interventions and environmental factors in disease.
- Human genomics is not the only part of genomics relevant to human health. The human genome interacts with those of a myriad other organisms, including plants, vectors and pathogens.
- Genomics is considered across all organisms, as relevant to public heath in human populations. In addition to genomics knowledge, we also considers technologies that make use of genomics knowledge.
- Genomics is distinct from genetics. While genetics is the study of heredity, genomics is defined as the study of genes and their functions, and related techniques.
- The main difference between genomics and genetics is that genetics scrutinizes the functioning and composition of the single gene where as genomics addresses all genes and their inter relationships in order to identify their combined influence on the growth and development of the organism.
What is the potential of genomics for public health?
- The role of human genomics research and related biotechnologies has the potential to achieve a number of public health goals, such as to reduce global health inequalities by providing developing countries with efficient, cost-effective and robust means of preventing, diagnosing and treating major diseases that burden their populations.
- It is a new and rapidly evolving branch of science and the full future role of genomics for the provision of health care is far from clear. However, it does offer the long-term possibility of providing new approaches to the prevention and management of many intractable diseases.
- Hence it is important to prepare society for the complexities of this new field, to ensure that its benefits are distributed fairly among the countries of the world, and that the well-tried and more conventional approaches to medical research and practice are not neglected while the medical potential of genomics is being explored.
Conclusion:
Since genome surveillance provides the sort of information that experts can use to devise an early response strategy, identify emerging viral strains, and undertake risk-based surveillance of key animal species, genomic technologies are likely to become mainstays of our arsenal against pathogens of the future.
Source: The Hindu
Mains Questions:
what do you mean by the term ‘genomics’ and ‘metagenomics’. Discuss its significance in health sector.